- Home
- Demonstrators
- Healthy Microbiome
The “Healthy Microbiome” demonstrator

Advancing diet-microbiome studies
The human microbiome, consisting of trillions of microorganisms residing in our bodies, plays a crucial role in digestion, immune function, and even mental health. By exploring the intricate relationship between our diet and our microbiome, we can learn how what we eat influences our gut microbiome and, in turn, impacts our overall health and well-being, paving the way for advancements in personalised nutrition and healthcare.
In the FNS-Cloud project, we developed a powerful platform to streamline and enhance the analysis of microbiomes in relation to diet. Our goal is to consolidate fragmented FNS-data from various sources into a seamless workflow, ultimately improving our understanding of the interaction between diet and the microbiome.
Our open-source online tools enable researchers to easily find and merge microbiome-related datasets stored in public repositories. With just a few clicks, users can access a wealth of valuable information, empowering them to conduct comprehensive analyses and download relevant data.
Discover the tools below 👇👇👇
FAIRSPACE
A FAIR data management platform
A FAIR data management platform
FAIRSPACE is an open-source research data management platform that adheres to the FAIR principles (Findability, Accessibility; Interoperability and Reusability). It was customised to become the FNS-Cloud metadata browser that allows microbiome and food data exploration within public resources. It brings datasets from different repositories together and links them at sample level.
Accessible from the FNS-Cloud Tools Catalogue.
Developed by The Hyve

Metadata submission form
A web-based tool to enhance microbiome studies
A web-based tool to enhance microbiome studies
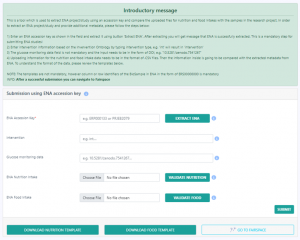
The form allows data curators to submit additional metadata (intervention, glucose monitoring data, and food or nutrition intake information) for existing public studies stored in the European Nucleotide Archive (ENA).
You can also submit metadata for microbiome studies not uploaded to the ENA database. This type of submission supports Ontology based input for many metadata fields.
Accessible from the FNS-Cloud Tools Catalogue.
Developed by ScaleFocus
Data analysis scripts
A Notebook for data pre-processing and exploration
A Notebook for data pre-processing and exploration
A series of scripts were developed to help answer the research questions. They can be used to analyse data and metadata using R, Python or Julia. A JupyterHub environment is integrated within Fairspace to allow users to run some of those predefined scripts for data pre-processing and exploratory analysis.
Accessible from the FAIRSPACE platform.
Developed by Quadram Institute
Target audience
Researchers in nutritional science, and anyone with an interest in diet and gut microbiome.
Use cases
To design the platform and demonstrate its potential use by researchers, the team worked around the three following research questions:
- Does a diet rich in plant bioactives affect our gut microbiome, and how?
— with the aim to demonstrate the user pathway and the handling of emerging metagenomics datasets and bioinformatic analytical pipelines. - Does our baseline gut microbiome define how species composition shifts upon dietary interventions?
— with the aim to demonstrate how to find, access and merge datasets from different diet & microbiome studies. - Does our gut microbiome contribute to our metabolic response to foods? Can we predict metabolic responses to foods using microbiome data? Is there more to food than nutrients?
— with the aim to demonstrate how users can access multiple datasets and a machine-learning prediction tool.
![]() Food Nutrition Security Cloud (FNS-Cloud) has received funding from the European Union’s Horizon 2020 Research and Innovation programme (H2020-EU.3.2.2.3. – A sustainable and competitive agri-food industry) under Grant Agreement No. 863059. Information and views set out across this website are those of the Consortium and do not necessarily reflect the official opinion or position of the European Union. Neither European Union institutions and bodies nor any person acting on their behalf may be held responsible for the use that may be made of the information contained herein.
Food Nutrition Security Cloud (FNS-Cloud) has received funding from the European Union’s Horizon 2020 Research and Innovation programme (H2020-EU.3.2.2.3. – A sustainable and competitive agri-food industry) under Grant Agreement No. 863059. Information and views set out across this website are those of the Consortium and do not necessarily reflect the official opinion or position of the European Union. Neither European Union institutions and bodies nor any person acting on their behalf may be held responsible for the use that may be made of the information contained herein.
